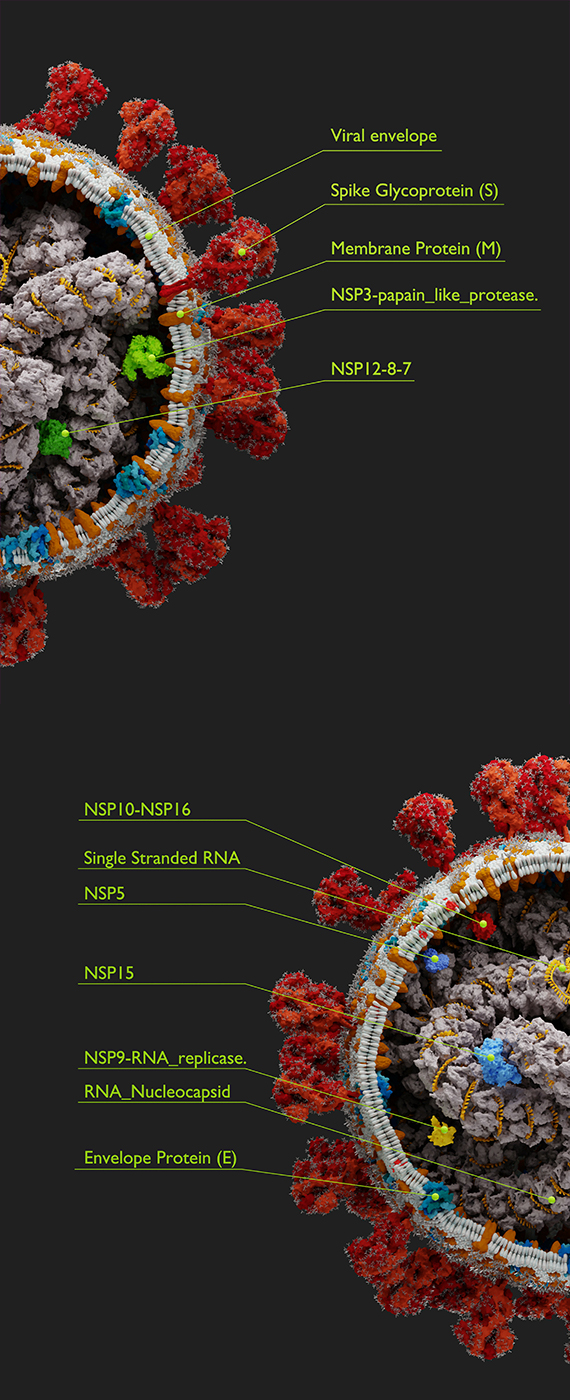
COVID-19 & SARS-CoV-2
Our lab's expertise in Data Science and Genomics have been put to use in COVID-19 research, including genome sequencing of SARS-CoV-2 genomes -- the virus responsible for the disease more commonly known as COVID-19. This work is currently ongoing with collaborators from Queen's University and Kingston Health Sciences Centre including Dr. Prameet Sheth, Dr. Calvin Sjaarda and others (see coauthors on papers linked below).
Phlogenomics of early-stage COVID-19 patients in Ontario, Canada
Our first research paper (published in Nature Scientific Reports) describes our sequencing of SARS-CoV-2 genomes using two different technologies, including a 'handheld' nanopore-based sequencer called the MinION by Oxford Nanopore. We analyzed SARS-CoV-2 genomes using evolutionary phylogenetics to identify sources of introduction and spread among the first COVID-19 cases in eastern Ontario. This work shows how portable sequencers and applied evolution can improve the public health response to emerging pandemics.
Fully reproducible code for this project is available on GitHub.
Temporal Dynamics and Evolution of SARS-CoV-2 in Ontario, Canada
Different genetic lineages (i.e. variants) have different mutations that may affect the rate of spread of COVID-19. In this study published in eSphere, we found variable infection rates in different lineages through time. Even in these early stages of the pandemic, variability in infection rates and the growing number of genetic lineages was cause for concern.
Metabolomics and Immune Response
Current projects (unpublished) track the immune response of COVID-19 patients and use chemistry of nasal swabs to identify biochemical changes in COVID-19 patients. These provide insights into the pathology of SARS-CoV-2, potential therapeutic targets, and the need for age-specific booster shots.